Nuclear organization
The genome self-organizes on the scale of 0.1-1 µm into a variety of chromatin subcompartments that assemble at certain sequences. Recent studies conclude that this process is driven by the unmixing of soluble factors from the nucleoplasm into protein droplets around chromatin by liquid-liquid phase separation (LLPS) to assembly transcriptionally silenced or active condensates. However, the local enrichment of proteins and RNAs can also be rationalized by stoichiometric binding to clustered binding sites in conjunction with long-range interactions between bound complexes that fold the nucleosome chain in 3D as depicted below.
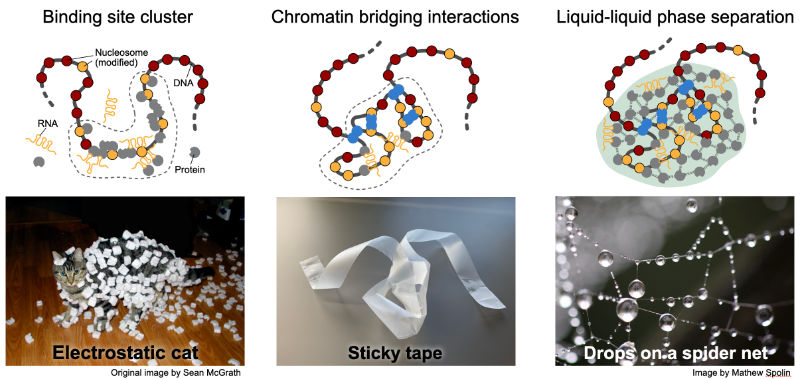 |
| Mechanisms of creating chromatin subcompartments that enrich specific genome associated activities. Clustering of binding sites, folding of the genome in space via long-range interactions and the non-stoichiometric accumulation of proteins and RNA around chromatin by phase separation. |
We study transcription activation at mouse pericentric heterochromatin, a reporter gene array and at endogenous gene cluster by both fluorescence microscopy and sequencing based methods. In these studies we elucidate the interplay of (i) direct protein binding to chromatin, (ii) the recruitment of additional factors by indirect interactions and the formation of liquid-like protein droplets, (iii) compaction/ decondensation of chromatin, and (iv) silencing/activation of transcription.
Key references
Trojanowski J, Frank L, Rademacher A, Mücke N, Grigaitis P, Rippe K (2022) Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol Cell 82, 1878-1893. doi: 10.1016/j.molcel.2022.04.017 | Abstract | Reprint | Article metrics
Rippe K (2022) Liquid-liquid phase separation in chromatin. Cold Spring Harb Perspect Biol 14, a040683. doi: 10.1101/cshperspect.a040683 | Abstract | Reprint | Article metrics
Erdel F, Rademacher A, Vlijm R, Tünnermann J, Schweigert E, Frank L, Yserantant K, Hummert J, Bauer C, Schumacher S, Weinmann R, Alwash AA, Normand C, Herten D-P, Engelhardt J, Rippe K (2020) Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol Cell 78, 236-249.e7. doi: 10.1016/j.molcel.2020.02.005 | Abstract | Reprint | Article metrics | Comment
