Chromatin Remodelers
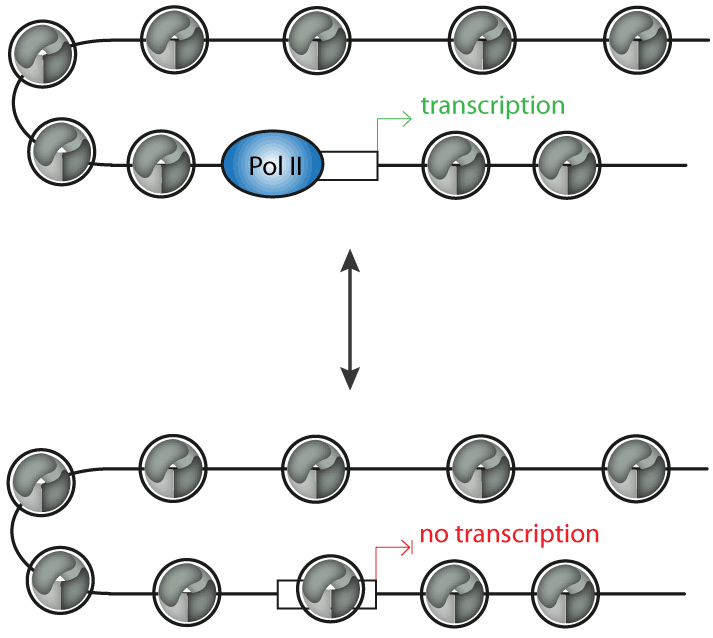
Chromatin remodelers are ATP-driven molecular machines that can alter nucleosome positions. This changes the accessibility of the DNA template in the context of chromatin, which is relevant for the regulation of transcription, DNA replication or repair (Figure 1).
Figure 1: Chromatin remodeling complexes can regulate transcription by controlling the position of promoter nucleosomes. Promoters that are free of nucleosomes can be bound by the transcription machinery and are permissive for transcription, whereas promoters that are occupied by nucleosomes do not allow transcription.
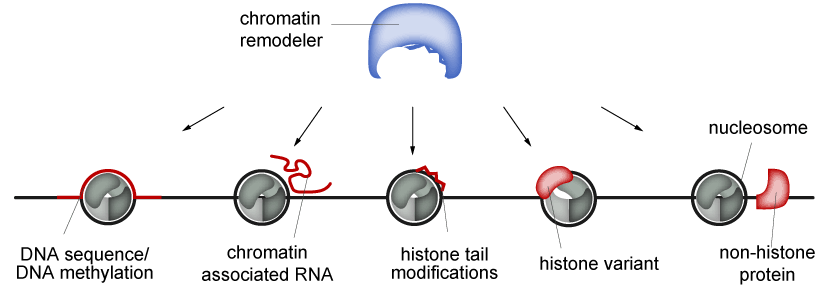
The catalytic subunits exist in different flavors and are highly abundant in the cell nucleus. The diversity is further increased by the formation of different chromatin remodeling complexes (CRCs) through association with non-catalytic subunits, which regulate both the catalytic activity and the targeting of CRCs. Identification of remodeling substrates is mediated by different cues, including histone or DNA modifications, histone variants, DNA sequence features and the length of the linker DNA (Figure 2).
Figure 2: Chromatin remodelers can be targeted by DNA, RNA, posttranslationally modified histone tails, histone variants or other chromatin-associated proteins.
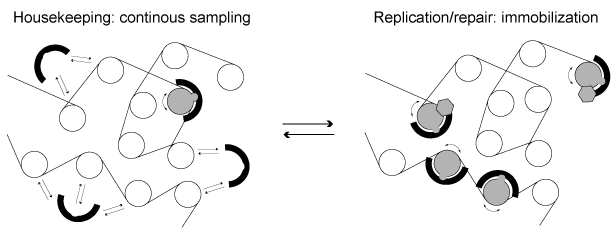
CRCs can also be recruited via DNA interacting proteins such as transcription factors or RNAs. However, little is known about the regulation of CRCs in living cells. In particular, it is unclear how specific substrates are selected and which signals are sufficient to trigger translocation. We address these questions by conducting mobility analyses in single living cells in combination with deep-sequencing based approaches. Since potential remodeling hotspots display reduced CRC mobility, fluctuation microscopy experiments are instructive for studying CRC targeting. Sequencing-based techniques as ChIP-Seq and MNase-Seq are used to obtain complementary information with higher resolution. Using this methodology, we have shown that human ISWI remodelers are only rarely active in unperturbed cells in G1 phase, suggesting that the amount of remodeling substrates is limited (Figure 3). In contrast, at replication foci in S phase or at DNA damage sites ISWIs become strongly immobilized.
Figure 3: ISWI remodelers diffuse through the nucleus most of the time and sample potential nucleosome substrates via transient binding interactions. In some special cases, such as at replication foci in S phase or at DNA damage sites, they bind chromatin more tightly and exert their activity.
References
- Erdel F, Rippe K. (2012).
Quantifying transient binding of ISWI chromatin remodelers in living cells by pixel-wise photobleaching profile evolution analysis.
PNAS 109(47):E3221-30. - Erdel F, Rippe K. (2011).
Chromatin remodeling in mammalian cells by ISWI type complexes – where, when and why?
FEBS J. 278(19):3608-18. - Erdel F, Krug J, Längst G, Rippe K. (2011).
Targeting chromatin remodelers: signals and search mechanisms.
Biochim Biophys Acta 1809(9):497-508. - Erdel F, Rippe K. (2011).
Binding kinetics of human ISWI chromatin-remodelers to DNA repair sites elucidate their target location mechanism.
Nucleus 2(2):105-12. - Erdel F, Schubert T, Marth C, Längst G, Rippe K. (2010).
Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites.
PNAS 107(46):19873-8.
People working on this project
Applied methods & techniques
Fluorescence Imaging
Single Cell Manipulations
Fluctuation Microscopy
Deep Sequencing
Fluctuation Microscopy
Deep Sequencing