Spatial Omics
Fluorescence microscopy and single cell sequencing approaches are integrated in spatially resolved omics methods to link molecular profiles of single cells in their tissue context with imaging based phenotypes. The spatial transcriptomics analysis is conducted on dedicated systems (Molecular Cartograpgy (Resolve Biosciences), Merscope (Vizgen), Xenium (10x Genomics). In several projects, we combine the spatial transcriptomics analysis with addtional readouts that are simultaneously acquired with an Andor Dragonfly spinning disk system at high-resolution together with the DAPI staining of nuclei as illustrated below. The images are registered with the spatial transcriptomics data via the DAPI signal, which is also used for segmentation of the nuclei. In this manner transcriptome analysis is combined with other modalities for an integrative analysis.
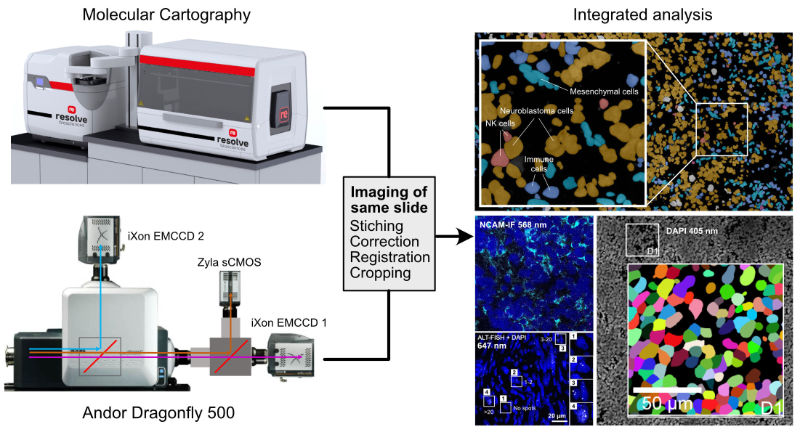 |
| Multi-modal imaging for spatially resolved transcriptomics analysis in combination with additional readouts. |
This approach is currently applied to resolve the transcriptome heterogeneity of tumor cells together with the activity of the alternative lengthening of telomeres pathway, immunostaining with specific tumor markers as well as genetic aberrations such as MYCN amplifications.
Key references
Ghasemi DR, Okonechnikov K, Rademacher A, Tirier S, Maass KK, Schumacher H, Sundheimer J, Statz B, Rifaioglu AS, Bauer K, Schumacher S, Bortolomeazzi M, Giangaspero F, Ernst KJ, Saez-Rodriguez J, Jones DTW, Kawauchi D, Mallm JP, Rippe K, Korshunov A, Pfister SM, Pajtler KW (2023) Compartments in medulloblastoma with extensive nodularity are connected through differentiation along the granular precursor lineage. Preprint bioRxiv 2022.09.02.506321. doi 10.1101/2022.09.02.506321 | Reprint | Article metrics
