Publications
Publications
2024
Seufert I, Gerosa I, Varamogianni-Mamatsi V, Vladimirova A, Sen E, Mantz S, Rademacher A, Schumacher S, Liakopoulos P, Kolovos P, Anders S, Mallm J-P, Papantonis A, Rippe K (2024) Two distinct chromatin modules regulate proinflammatory gene expression. bioRxiv, 2024.2008.2003.606159. doi: 10.1101/2024.08.03.606159 | Abstract | Reprint | Article metrics
Rademacher A, Huseynov A, Bortolomeazzi M, Wille SJ, Schumacher S, Sant P, Keitel D, Okonechnikov K, Ghasemi D, Pajtler K, Mallm J-P, Rippe K (2024) Comparison of spatial transcriptomics technologies used for tumor cryosections. bioRxiv, 2024.2004.2003.586404. doi: 10.1101/2024.04.03.586404 | Abstract | Reprint | Article metrics
Okonechnikov K, Joshi P, Koerber V, Rademacher A, Bortolomeazzi M, Mallm J-P, da Silva PBG, Statz B, Sepp M, Sarropoulos I, Yamada-Saito T, Vaillant J, Wittmann A, Schramm K, Blattner-Johnson M, Fiesel P, Jones B, Milde T, Pajtler K, van Tilburg CM, Witt O, Bochennek K, Weber KJ, Nonnenmacher L, Reimann C, Schueller U, Mynarek M, Rutkowski S, Jones DTW, Korshunov A, Rippe K, Westermann F, Thongjuea S, Hoefer T, Kaessmann H, Kutscher LM, Pfister SM (2024) Medulloblastoma oncogene aberrations are not involved in tumor initiation, but essential for disease progression and therapy resistance. bioRxiv, 2024.2002.2009.579690. doi: 10.1101/2024.02.09.579690 | Abstract | Reprint | Article metrics
Joshi P, Stelzer T, Okonechnikov K, Sarropoulos I, Sepp M, Pour-Jamnani MV, Rademacher A, Yamada-Saito T, Schneider C, Schmidt J, Schäfer P, Leiss K, Bortolomeazzi M, Mallm J-P, da Silva PBG, Statz B, Wittmann A, Schramm K, Blattner-Johnson M, Fiesel P, Jones B, Milde T, Pajtler K, van Tilburg CM, Witt O, Rippe K, Korshunov A, Jones DTW, Hovestadt V, Northcott PA, Thongjuea S, Jäger N, Kaessmann H, Pfister SM, Kutscher LM (2024) Gene regulatory network landscape of Group 3/4 medulloblastoma. bioRxiv,2024.2002.2009.579680. doi: 10.1101/2024.02.09.579680 | Abstract | Reprint | Article metrics
Lutz R, Grünschläger F, Simon M, Awwad MHS, Bauer M, Yousefian S, Beumer N, Jopp-Saile L, Sedlmeier A, Solé-Boldo L, Avanesyan B, Vonficht D, Stelmach P, Steinbuss G, Boch T, Steiger S, Baertsch MA, Prokoph N, Rippe K, Durie BGM, Wickenhauser C, Trumpp A, Müller-Tidow C, Hübschmann D, Weinhold N, Raab MS, Brors B, Goldschmidt H, Imbusch CD, Hundemer M, Haas S (2024) Multiple myeloma long-term survivors display sustained immune alterations decades after first line therapy. Nat Commun, accepted in principle.
Cirrincione AM, Poos AM, Ziccheddu B, Kaddoura M, Bärtsch M-A, Maclachlan K, Chojnacka M, Diamond B, John L, Reichert P, Huhn S, Blaney P, Gagler D, Rippe K, Zhang Y, Dogan A, Lesokhin AM, Davies F, Goldschmidt H, Fenk R, Weisel KC, Mai EK, Korde N, Morgan GJ, Usmani S, Landgren O, Raab MS, Weinhold N, Maura F (2024) The biological and clinical impact of deletions before and after large chromosomal gains in multiple myeloma. Blood 144, 771-783. doi: 10.1182/blood.2024024299 | Abstract | Reprint | Article metrics
Roessner PM, Seufert I, Chapaprieta V, Jayabalan R, Briesch H, Massoni-Badosa R, Boskovic P, Beckendorff J, Roider T, Arseni L, Coelho M, Chakraborty S, Vaca AM, Sivina M, Muckenhuber M, Rodriguez-Rodriguez S, Bonato A, Herbst SA, Zapatka M, Sun C, Kretzmer H, Naake T, Bruch P-M, Czernilofsky F, Hacken ET, Schneider M, Helm D, Yosifov DY, Kauer J, Danilov AV, Bewarder M, Heyne K, Schneider C, Stilgenbauer S, Wiestner A, Mallm JP, Burger JA, Efremov DG, Lichter P, Dietrich S, Martin-Subero JI, Rippe K, Seiffert M (2024) T-bet suppresses proliferation of malignant B cells in chronic lymphocytic leukemia. Blood 144, 510-524. doi: 10.1182/blood.2023021990 | Abstract | Reprint | Article metrics
Gilbertson RJ, Behjati S, Bottcher AL, Bronner ME, Burridge M, Clausing H, Clifford H, Danaher T, Donovan LK, Drost J, Eggermont AMM, Emerson C, Flores MG, Hamerlik P, Jabado N, Jones A, Kaessmann H, Kleinman CL, Kool M, Kutscher LM, Lindberg G, Linnane E, Marioni JC, Maris JM, Monje M, Macaskill A, Niederer S, Northcott PA, Peeters E, Plieger-van Solkema W, Preussner L, Rios AC, Rippe K, Sandford P, Sgourakis NG, Shlien A, Smith P, Straathof K, Sullivan PJ, Suva ML, Taylor MD, Thompson E, Vento-Tormo R, Wainwright BJ, Wechsler-Reya RJ, Westermann F, Winslade S, Al-Lazikani B, Pfister SM (2024) The Virtual Child. Cancer Discov 14, 663-668. doi: 10.1158/2159-8290.CD-23-1500 | Abstract | Reprint | Article metrics
Bundschuh C, Weidner N, Klein J, Rausch T, Azevedo N, Telzerow A, Mallm JP, Kim H, Steiger S, Seufert I, Borner K, Bauer K, Hubschmann D, Jost KL, Parthe S, Schnitzler P, Boutros M, Rippe K, Muller B, Bartenschlager R, Krausslich HG, Benes V (2024) Evolution of SARS-CoV-2 in the Rhine-Neckar/Heidelberg Region 01/2021 - 07/2023. Infect Genet Evol, 105577. doi: 10.1016/j.meegid.2024.105577 | Abstract | Reprint | Article metrics
Ghasemi DR, Okonechnikov K, Rademacher A, Tirier S, Maass KK, Schumacher H, Sundheimer J, Statz B, Rifaioglu AS, Bauer K, Schumacher S, Bortolomeazzi M, Giangaspero F, Ernst KJ, Saez-Rodriguez J, Jones DTW, Kawauchi D, Mallm JP, Rippe K, Korshunov A, Pfister SM, Pajtler KW (2024) Compartments in medulloblastoma with extensive nodularity are connected through differentiation along the granular precursor lineage. Nat Commun 15, 269. doi: 10.1038/s41467-023-44117-x | Abstract | Reprint | Article metrics | Press release
Wagner TR, Kehl N, Steiger S, Kilian M, Foster K, Boschert T, Hernandez GM, Abelin J, Lindner K, Sester LS, Frenking J, Schönfelder B, Galas-Filipowicz D, Fitzsimons E, Green EW, Schmidt P, Uhrig S, Bunse L, Chain B, Goldschmidt H, Weinhold N, Fröhling S, Müller-Tidow C, Carr SA, Yong K, Rippe K, Raab MS, Platten M, Eichmüller SB, Friedrich MJ. Tumor reactive T cells reside in the bone marrow and are associated with clinical response to hematological malignancies. Blood 142 (Supplement 1), 4074-4074 (ASH 2023 Abstract). doi: 10.1182/blood-2023-189779 | Abstract | Reprint
Selected recent publications 2019-2023 (more selected publications)
Piroeva KV, McDonald C, Xanthopoulos C, Fox C, Clarkson CT, Mallm J-P, Vainshtein Y, Ruje L, Klett LC, Stilgenbauer S, Mertens D, Kostareli E*, Rippe K*, Teif VB* (2023) Nucleosome repositioning in chronic lymphocytic leukaemia. Genome Res 33, 1649-1661. *corresponding authors. doi: 10.1101/gr.277298.122 | Abstract | Reprint | Article metrics
Poos AM, Prokoph N, Przybilla MJ, Mallm J-P, Steiger S, Seufert I, John L, Tirier SM, Bauer K, Baumann A, Munawar U, Rasche L, Kortuem M, Giesen N, Huhn S, Reichert P, Mueller-Tidow C, Goldschmidt H, Stegle O, Raab MS, Rippe K*, Weinhold N* (2023) Resolving therapy resistance mechanisms in multiple myeloma by multiomics subclone analysis. Blood 142, 1633-1646. *corresponding authors. doi: 10.1182/blood.2023019758 | Abstract | Reprint | Article metrics | Press release (German)
Trojanowski J, Frank L, Rademacher A, Mücke N, Grigaitis P, Rippe K (2022) Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol Cell 82, 1878-1893. doi: 10.1016/j.molcel.2022.04.017 | Abstract | Reprint | Article metrics
Frank L, Rademacher A, Mücke N, Tirier SM, Koeleman E, Knotz C, Schumacher S, Stainczyk SA, Westermann F, Fröhling S, Chudasama P, Rippe K (2022) ALT-FISH quantifies alternative lengthening of telomeres activity by imaging of single-stranded repeats. Nucleic Acids Res 50, e61. doi: 10.1093/nar/gkac113 | Abstract | Reprint | Article metrics
Tirier SM, Mallm J-P, Steiger S, Poos AM, Awwad M, Giesen N, Casiraghi N, Susak H, Bauer K, Baumann A, John L, Seckinger A, Hose D, Müller-Tidow C, Goldschmidt H, Stegle O, Hundemer M, Weinhold N, Raab MS, Rippe K (2021) Subclone specific microenvironmental impact and drug response in refractory multiple myeloma revealed by single cell transcriptomics. Nat Commun 12, 6960. doi: 10.1038/s41467-021-26951-z | Abstract | Reprint | Article metrics
Erdel F, Rademacher A, Vlijm R, Tünnermann J, Schweigert E, Frank L, Yserantant K, Hummert J, Bauer C, Schumacher S, Weinmann R, Alwash AA, Normand C, Herten D-P, Engelhardt J, Rippe K (2020) Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol Cell 78, 236-249.e7. doi: 10.1016/j.molcel.2020.02.005 | Abstract | Reprint | Article metrics | Comment
Mallm JP, Windisch P, Biran A, Schumacher S, Glass R, Herold-Mende C, Meshorer E, Barbus M, Rippe K (2020) Glioblastoma initiating cells are sensitive to histone demethylase inhibition due to loss of DNA repair capacity. Intl J Cancer 146, 1281-1292. doi: 10.1002/ijc.32649 | Abstract | Reprint | Article metrics
Mallm JP, Iskar M, Ishaque N, Klett LC, Kugler SJ, Muino JM, Teif VB, Poos AM, Großmann S, Erdel F, Tavernari D, Koser SD, Schumacher S, Brors B, König R, Remondini D, Vingron M, Stilgenbauer S, Lichter P, Zapatka M, Mertens D & Rippe K (2019) Linking aberrant chromatin features in chronic lymphocytic leukemia to deregulated transcription factor networks. Mol Syst Biol 15, e8339. doi: 10.15252/msb.20188339 | Abstract | Reprint | Appendix | Additional data | Article metrics
All publications 1989-2023
All publications Karsten Rippe, h-index 74 (Google Scholar)
- Research articles in peer reviewed journals (158 articles)
- Reviews and book chapters (peer reviewed) (36 articles)
- Selected publications
- Other publications (6 articles)
- PhD theses (23), diploma/master theses (18) and patents (2)
Publications and bibliometric data of Karsten Rippe from databases
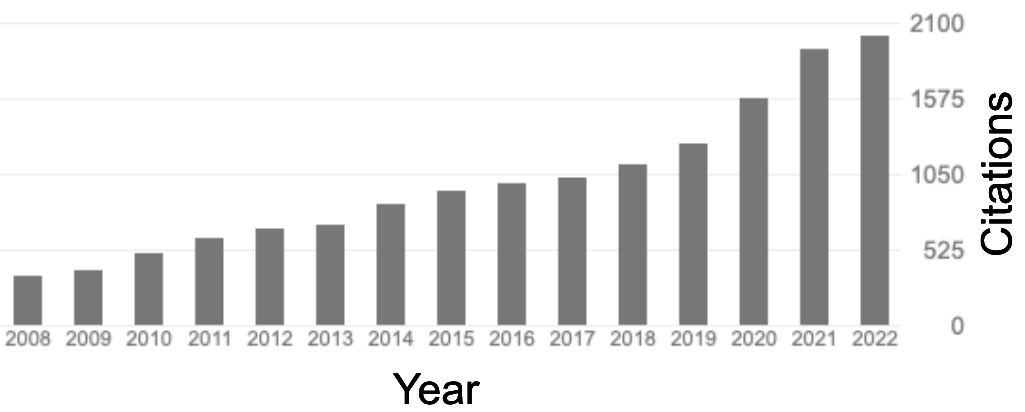 |
Yearly citation numbers of Karsten Rippe from 2008 to 2022 from Google Scholar. Total number citations: 18065 h-index: 71 i10 index: 162 |
