Chromatin in Leukemia
Background
Changes in the epigenome cause aberrant gene expression patterns that are associated with cancer development. Several epigenetic drugs have been developed that have the potential to reprogram the epigenome and showed good clinical responses.
We investigate the network of epigenetic modifications, non-coding RNAs, nucleosome positioning and transcription factor activity to determine the molecular processes that result and maintain cancer, specifically in leukemia.
Work of the group
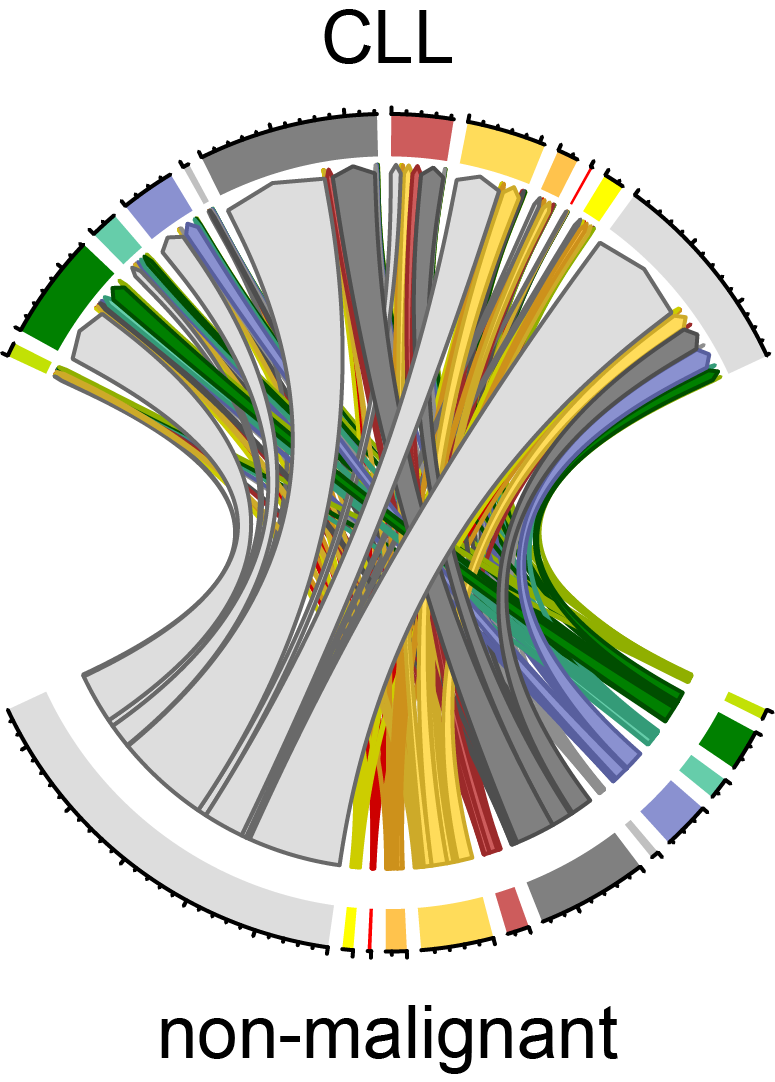
Chronic lymphocytic leukemia (CLL)
Within the PRECiSe consortium we set out to dissect the epigenetic networks associated with chronic lymphocytic leukemia (CLL) to develop novel diagnostic and therapeutic approaches for the disease. Furthermore, we are identifying novel epigenetic biomarkers by understanding how BCR signaling impacts on the deregulated transcriptional and epigenetic landscape in CLL patient cells in the BMBF funded PRECiSe project. We aim to overcome chemoresistance by understanding the molecular mode of action via its imprint on the epigenetic networks in CLL. Eventually, we will exploit the opportunity to identify epigenetic signatures that predict response to first-line treatment in asymptomatic patients. For further information please visit our PRECiSe website at www.CancerEpiSys.org
 |
|
Acute myeloid leukemia (AML)
Within the DFG Research Unit 2674 Aging-related epigenetic remodeling in acute myeloid leukemia we collaborate with research groups in Heidelberg, Ulm and Freiburg to uncover age-related epigenetic remodeling in acute myeloid leukemia (AML) by single-cell sequencing methods. Our group will focus on response to treatment in the context of tumor heterogeneity in AML cases that have a mutation of IDH1.
Multiple myeloma (MM)
In a collaboration with Prof. Dr. Hartmut Goldschmidt and Dr. Marc-Steffen Raab from the University Hospital Heidelberg we are investigating both interindividual differences and intrapatient heterogeneity in the response to an epigenetically active drug in multiple myeloma. Furthermore, we study resistance mechanisms in relapsed-refractory multiple myeloma by investigating the change in tumor composition during resistance development by single-cell sequencing. This work is conducted in the Heidelberg Center for Personalized Oncology (DKFZ-HIPO) program within HIPO2 project K08 and HIPO2 project K43.
Selected references
Bhattacharya N, Reichenzeller M, Caudron-Herger M, Haebe S, Brady N, Diener S, Nothing M, Döhner H, Stilgenbauer S, Rippe K & Mertens D (2015). Loss of cooperativity of secreted CD40L and increased dose-response to IL4 on CLL cell viability correlates with enhanced activation of NF-kB and STAT6. Int J Cancer 136, 65-73. doi: 10.1002/ijc.28974 | Abstract | Reprint (3.0 MB) | Article metrics
Chaturvedi A, Herbst L, Pusch S, Klett L, Goparaju R, Stichel D, Kaulfuss S, Panknin O, Zimmermann K, Toschi L, Neuhaus R, Haegebarth, A, Rehwinkel H, Hess-Stumpp H, Bauser M, Bochtler T, Struys EA, Sharma A, Bakkali A, Geffers R, Araujo-Cruz MM, Thol F, Gabdoulline R, Ganser A, Ho AD, von Deimling A, Rippe K, Heuser M & Kraemer A (2017). Pan-mutant IDH1 inhibitor BAY1436032 is highly effective against human IDH1 mutant acute myeloid leukemia in vivo. Leukemia 31, 2020-2028. doi: 10.1038/leu.2017.46 | Abstract | Reprint (2.2 MB) | Article metrics.
Delacher M, Imbusch CD, Weichenhan D, Breiling A, Hotz-Wagenblatt A, Trager U, Hofer AC, Kagebein D, Wang Q, Frauhammer F, Mallm JP, Bauer K, Herrmann C, Lang PA, Brors B, Plass C & Feuerer M (2017). Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat Immunol 18, 1160-1172. doi: 10.1038/ni.3799 | Abstract | Article metrics.
Teif VB, Mallm JP, Sharma T, Welch DBM, Rippe K, Eils R, Langowski J, Olins AL & Olins DE (2017). Nucleosome repositioning during differentiation of a human myeloid leukemia cell line. Nucleus 8, 188-204. doi: 10.1080/19491034.2017.1295201 | Abstract | Reprint (1.8 MB) | Article metrics.
Related research
Related methods
Links
- PRECiSe consortium: www.CancerEpiSys.org
- DFG Research Unit 2674 Aging-related epigenetic remodeling in acute myeloid leukemia
- HIPO2 project K08: Personalized treatment strategies and resistance mechanisms in relapsed-refractory multiple myeloma
- HIPO2 project K43: Dissecting the treatment-associated evolution of the clonal heterogeneity of tumor cells and their microenvironment in multiple myeloma
