Publications
Publications
2025
Seufert I, Gerosa I, Varamogianni-Mamatsi V, Vladimirova A, Sen E, Mantz S, Rademacher A, Schumacher S, Liakopoulos P, Kolovos P, Anders S, Mallm JP, Papantonis A, Rippe K (2025) Two distinct chromatin modules regulate proinflammatory gene expression. Nat Cell Biol, in press. bioRxiv preprint doi: 10.1101/2024.08.03.606159 | Abstract | Reprint | Article metrics
Alberti S, Arosio P, Best R, Boeynaems S, Cai D, Collepardo-Guevara R, Dignon G, Dimova R, Elbaum-Garfinkle S, Fawzi N, Fuxreiter M, Gladfelter A, Honigmann A, Jain A, Joseph J, Knowles T, Lasker K, Lemke E, Lindorff-Larsen K, Lipowsky R, Mittal J, Mukhopadhyay S, Myong S, Pappu R, Rippe K, Shelkovnikova T, Vecchiarelli A, Wegmann S, Zhang H, Zhang M, Zubieta C, Zweckstetter M, Dormann D, Mittag T (2025) Current practices in the study of biomolecular condensates: a community comment. Nat Commun 16, 7730. doi: 10.1038/s41467-025-62055-8 | Abstract | Reprint | Article metrics
Staehle HF, Staehle AM, Köllerer C, Schulze J, Mallm J-P, Pozdnyakova O, Rippe K, Brors B, Lipka D, Schüle R, Pahl HL, Jutzi JS (2025) Lysine-specific demethylase 1 regulates hematopoietic stem cell expansion and myeloid cell differentiation. Cell Death & Disease 16, 619. doi: 10.1038/s41419-025-07951-z | Abstract | Reprint | Article metrics
Rippe K, Papantonis A (2025) RNA polymerase II ranscription compartments – from factories to condensates. Nat Rev Genet, published online 19 June 2025. doi: https://doi.org/10.1038/s41576-025-00859-6 | Abstract | Reprint | Article metrics
Rademacher A, Huseynov A, Bortolomeazzi M, Wille SJ, Schumacher S, Sant P, Keitel D, Okonechnikov K, Ghasemi D, Pajtler K, Mallm JP, Rippe K (2025) Comparison of spatial transcriptomics technologies used for tumor cryosections. Genome Biol 26, 176. doi: 10.1101/2024.04.03.586404 | Abstract | Reprint | Article metrics
Okonechnikov K, Joshi P, Koerber V, Rademacher A, Bortolomeazzi M, Mallm J-P, da Silva PBG, Statz B, Sepp M, Sarropoulos I, Yamada-Saito T, Vaillant J, Wittmann A, Schramm K, Blattner-Johnson M, Fiesel P, Jones B, Milde T, Pajtler K, van Tilburg CM, Witt O, Bochennek K, Weber KJ, Nonnenmacher L, Reimann C, Schueller U, Mynarek M, Rutkowski S, Jones DTW, Korshunov A, Rippe K, Westermann F, Thongjuea S, Hoefer T, Kaessmann H, Kutscher LM, Pfister SM (2025) Medulloblastoma oncogene aberrations are not involved in tumor initiation, but essential for disease progression and therapy resistance. Nature 642, 1062-1072. doi: 10.1038/s41586-025-08973-5 | Abstract | Reprint | Article metrics | Press release
Seufert I, Vargas C, Wille SJ, Rippe K (2025) Deregulated enhancer-promoter communication in cancer through altered nuclear architecture. Int J Cancer, published online 12 April 2025. doi: 10.1002/ijc.35424 | Abstract | Reprint | Article metrics
Lutz R, Poos AM, Sole-Boldo L, John L, Wagner J, Prokoph N, Baertsch MA, Vonficht D, Palit S, Brobeil A, Mechtersheimer G, Hildenbrand N, Hemmer S, Steiger S, Horn S, Pepke W, Spranz DM, Rehnitz C, Sant P, Mallm JP, Friedrich MJ, Reichert P, Huhn S, Trumpp A, Rippe K, Haghverdi L, Frohling S, Muller-Tidow C, Hubschmann D, Goldschmidt H, Willimsky G, Sauer S, Raab MS, Haas S, Weinhold N (2025) Bone marrow breakout lesions act as key sites for tumor-immune cell diversification in multiple myeloma. Sci Immunol 10, eadp6667. doi: 10.1126/sciimmunol.adp6667 | Abstract | Reprint | Article metrics | Press release
Preprints
Wagner TR, Kehl N, Steiger S, Boschert T, Kilian M, Foster K, Hernandez GM, Ctortecka C, Michel J, Schach A, Zoller J, Metzler A, Onken R, Schönfelder B, Renders S, Torres CM, Sester LS, Frenking JH, Werner F, Osen W, Lindner K, Sen E, Schumacher S, Galas-Filipowicz D, Fitzsimons E, Green EW, Schmidt P, Lindner JM, Uhrig S, Bunse L, Chain B, Goldschmidt H, Weinhold N, Fröhling S, Trumpp A, Abelin JG, Carr SA, Yong K, Müller-Tidow C, Rippe K, Raab MS, Platten M, Eichmüller SB, Friedrich MJ (2025) Conserved programs and specificities of T cells targeting hematological malignancies. bioRxiv: 2025.2008.2007.668919. doi: 10.1101/2025.08.07.668919 | Abstract | Reprint | Article metrics
Joshi P, Stelzer T, Okonechnikov K, Sarropoulos I, Sepp M, Pour-Jamnani MV, Rademacher A, Yamada-Saito T, Schneider C, Schmidt J, Schäfer P, Leiss K, Bortolomeazzi M, Mallm J-P, da Silva PBG, Statz B, Wittmann A, Schramm K, Blattner-Johnson M, Fiesel P, Jones B, Milde T, Pajtler K, van Tilburg CM, Witt O, Rippe K, Korshunov A, Jones DTW, Hovestadt V, Northcott PA, Thongjuea S, Jäger N, Kaessmann H, Pfister SM, Kutscher LM (2024) Gene regulatory network landscape of Group 3/4 medulloblastoma. bioRxiv,2024.2002.2009.579680. doi: 10.1101/2024.02.09.579680 | Abstract | Reprint | Article metrics
Weinmann R, Udupa A, Nickels JF, Frank L, Knotz C, Rippe K (2024) HP1 binding creates a local barrier against transcription activation and persists during chromatin decondensation. bioRiv, 2024.2012.2009.627308. doi: 2024.2012.2009.627308 | Abstract | Reprint | Article metrics
Selected recent publications 2019-2023 (more selected publications)
Piroeva KV, McDonald C, Xanthopoulos C, Fox C, Clarkson CT, Mallm J-P, Vainshtein Y, Ruje L, Klett LC, Stilgenbauer S, Mertens D, Kostareli E*, Rippe K*, Teif VB* (2023) Nucleosome repositioning in chronic lymphocytic leukaemia. Genome Res 33, 1649-1661. *corresponding authors. doi: 10.1101/gr.277298.122 | Abstract | Reprint | Article metrics
Poos AM, Prokoph N, Przybilla MJ, Mallm J-P, Steiger S, Seufert I, John L, Tirier SM, Bauer K, Baumann A, Munawar U, Rasche L, Kortuem M, Giesen N, Huhn S, Reichert P, Mueller-Tidow C, Goldschmidt H, Stegle O, Raab MS, Rippe K*, Weinhold N* (2023) Resolving therapy resistance mechanisms in multiple myeloma by multiomics subclone analysis. Blood 142, 1633-1646. *corresponding authors. doi: 10.1182/blood.2023019758 | Abstract | Reprint | Article metrics | Press release (German)
Trojanowski J, Frank L, Rademacher A, Mücke N, Grigaitis P, Rippe K (2022) Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol Cell 82, 1878-1893. doi: 10.1016/j.molcel.2022.04.017 | Abstract | Reprint | Article metrics
Frank L, Rademacher A, Mücke N, Tirier SM, Koeleman E, Knotz C, Schumacher S, Stainczyk SA, Westermann F, Fröhling S, Chudasama P, Rippe K (2022) ALT-FISH quantifies alternative lengthening of telomeres activity by imaging of single-stranded repeats. Nucleic Acids Res 50, e61. doi: 10.1093/nar/gkac113 | Abstract | Reprint | Article metrics
Tirier SM, Mallm J-P, Steiger S, Poos AM, Awwad M, Giesen N, Casiraghi N, Susak H, Bauer K, Baumann A, John L, Seckinger A, Hose D, Müller-Tidow C, Goldschmidt H, Stegle O, Hundemer M, Weinhold N, Raab MS, Rippe K (2021) Subclone specific microenvironmental impact and drug response in refractory multiple myeloma revealed by single cell transcriptomics. Nat Commun 12, 6960. doi: 10.1038/s41467-021-26951-z | Abstract | Reprint | Article metrics
Erdel F, Rademacher A, Vlijm R, Tünnermann J, Schweigert E, Frank L, Yserantant K, Hummert J, Bauer C, Schumacher S, Weinmann R, Alwash AA, Normand C, Herten D-P, Engelhardt J, Rippe K (2020) Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol Cell 78, 236-249.e7. doi: 10.1016/j.molcel.2020.02.005 | Abstract | Reprint | Article metrics | Comment
Mallm JP, Windisch P, Biran A, Schumacher S, Glass R, Herold-Mende C, Meshorer E, Barbus M, Rippe K (2020) Glioblastoma initiating cells are sensitive to histone demethylase inhibition due to loss of DNA repair capacity. Intl J Cancer 146, 1281-1292. doi: 10.1002/ijc.32649 | Abstract | Reprint | Article metrics
Mallm JP, Iskar M, Ishaque N, Klett LC, Kugler SJ, Muino JM, Teif VB, Poos AM, Großmann S, Erdel F, Tavernari D, Koser SD, Schumacher S, Brors B, König R, Remondini D, Vingron M, Stilgenbauer S, Lichter P, Zapatka M, Mertens D & Rippe K (2019) Linking aberrant chromatin features in chronic lymphocytic leukemia to deregulated transcription factor networks. Mol Syst Biol 15, e8339. doi: 10.15252/msb.20188339 | Abstract | Reprint | Appendix | Additional data | Article metrics
All publications 1989-2024
All publications Karsten Rippe, h-index 78 (Google Scholar)
- Research articles in peer reviewed journals (164 articles)
- Reviews and book chapters (peer reviewed) (36 articles)
- Selected publications
- Other publications (6 articles)
- PhD theses (25), Master's/Diploma theses (19) and patents (2)
Publications and bibliometric data of Karsten Rippe from databases
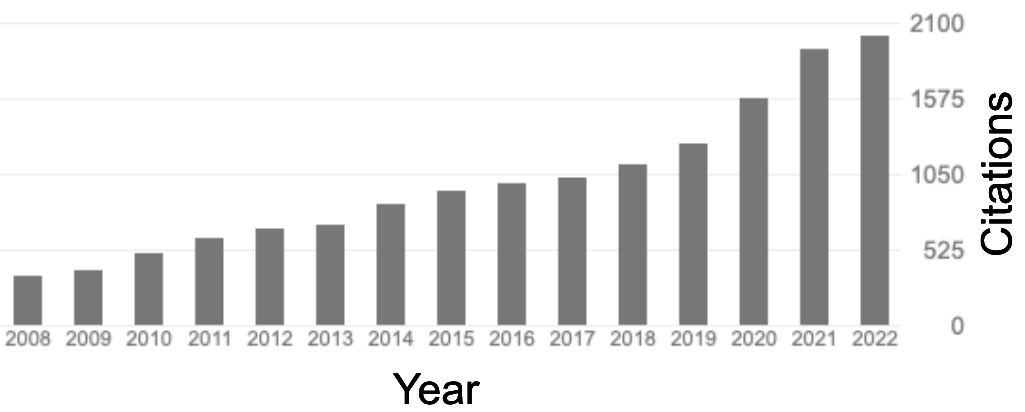 |
Yearly citation numbers of Karsten Rippe from 2008 to 2022 from Google Scholar. Total number citations: 18065 h-index: 71 i10 index: 162 |
