Telomere maintenance
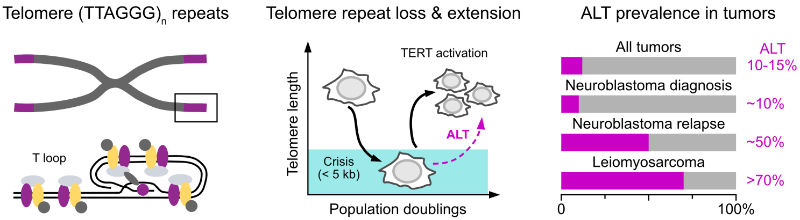
Telomeres are specialized structures at the ends of linear chromosomes. Their maintenance is essential for the unlimited proliferation of cells due to end-replication problem. Progressive telomere shortening in somatic cells can lead to the induction of senescence or apoptosis, thus acting as a barrier to unlimited proliferation. Therefore, maintaining telomere length is a crucial feature of cancers. To elongate their telomeres, most cancer cells reactivate telomerase, an enzyme which is normally only active in stem cells. However, in 10-15% of cancers a telomerase-independent pathway called alternative lengthening of telomeres (ALT) is activated. It exploits aberrant DNA repair and recombination at telomeres to counteract replicative senescence and to provide cancer cells with an unlimited proliferation potential. In tumors like relpased neuroblastoma and leiomyosarcoma, ALT occurs in around half of the patients and is linked to high-risk cases and poor outcome.
 |
| Establishing a telomere maintenance mechanism. |
ALT activity is rarely evaluated in clinical diagnostics, and treatments that specifically target ALT are not available. A major challenge for exploring ALT-specific vulnerabilities is the intra-tumor heterogeneity, which makes it difficult to reveal links between ALT activity and specific cellular phenotypes from the bulk analysis of molecular features that is conducted with current methods. This shortcoming is addressed in our work. We dissect how ALT activity is linked to cancer cell proliferation capacity and devise strategies to exploit the ALT mechanism for patient stratification and diagnosis as well as to identify specific vulnerabilities of ALT tumors.
Key references
Frank L, Rademacher A, Mücke N, Tirier SM, Koeleman E, Knotz C, Schumacher S, Stainczyk SA, Westermann F, Fröhling S, Chudasama P, Rippe K (2022) ALT-FISH quantifies alternative lengthening of telomeres activity by imaging of single-stranded repeats. Nucleic Acids Res, gkac113. doi: 10.1093/nar/gkac113 | Abstract | Reprint | Article metrics
Osterwald S, Deeg KI, Chung I, Parisotto D, Wörz S, Rohr K, Erfle H & Rippe K (2015). PML induces compaction, partial TRF2 depletion and DNA damage signaling at telomeres and promotes alternative lengthening of telomeres. J Cell Sci 128, 1887-1900. doi: 10.1242/jcs.148296 | Abstract | Reprint (4.3 MB) | Article metrics
Chung I, Leonhardt H & Rippe K (2011). De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J Cell Sci 124, 3603-3618. doi: 10.1242/jcs.084681 | Abstract | Reprint (9.4 MB) | Comment | Article metrics
